DNA
DNA
Read here about DNA considered as a tensegrity structure, and the use of DNA in the construction of the world's tinniest tensegrities.
Overview
DNA, or deoxyribonucleic acid, is a nucleic acid that contains the genetic instructions used in the development and functioning of all known living organisms and some viruses. The main role of DNA molecules is the long-term storage of information. DNA is often compared to a set of blueprints, like a recipe or a code, since it contains the instructions needed to construct other components of cells, such as proteins and RNA molecules.
Chemically, DNA consists of two long polymers of simple units called nucleotides, with backbones made of sugars and phosphate groups joined by ester bonds. These two strands run in opposite directions to each other and are therefore anti-parallel. Attached to each sugar is one of four types of molecules called bases. It is the sequence of these four bases along the backbone that encodes information. This information is read using the genetic code, which specifies the sequence of the amino acids within proteins. The code is read by copying stretches of DNA into the related nucleic acid RNA, in a process called transcription.
The structure of DNA makes it amenable to nano-construction techniques.
DNA as Compression members in Tensegrity Structures
Prof. "Ned" Nadrian C. Seeman constructs stick figures of DNA. His lab attaches specific sticky ends to DNA branched junctions, whose edges are double-stranded DNA. This approach has already been used to assemble a cube, a truncated octahedron , nanomechanical devices and 2-D crystals and 3-D crystals from DNA. This method of construction was originally stimulated by Seeman's desire to characterize Holliday junctions. Holliday junctions are four-arm branched DNA molecules that are found to be structural intermediates in genetic recombination. In the last few years, the symmetry, crossover topology and sequence-dependent thermodynamics of Holliday junctions have been characterized and analyzed.
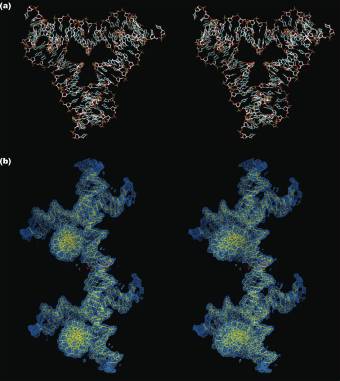
Molecular Structure of the 3 Strut Tensegrity Triangle Constructed from DNA
The stereoscopic images below show the molecular structure of the tensegrity triangle (a) and the connection between two of them through a sticky end (b).
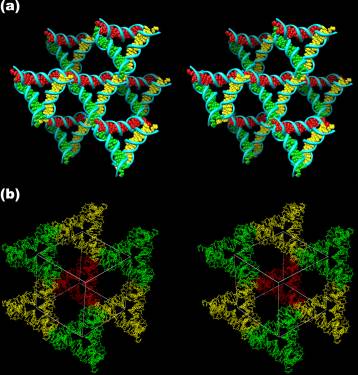
The stereoscopic images below the surroundings of one triangle (a) and the rhombohedron flanked by eight of the triangles (b). The red triangle is at the back of the rhombohedron, the three yellow triangles are in a plane closer to the viewer, and the three green triangles are in a plane yet closer to the viewer. The eighth red triangle which would be yet closer to the viewer has been omitted for clarity.
Other Polyhedra Created from DNA by Seeman's Team
This representation of a DNA cube shows that it contains six different cyclic strands. Their backbones are shown in red (front), green (right), yellow (back), magenta (left), cyan (top) and dark blue (bottom). Each nucleotide is represented by a single colored dot for the backbone and a single white dot representing the base. Note that the helix axes of the molecule have the connectivity of a cube. However, the strands are linked to each other twice on every edge. Therefore, this molecule is a hexacatenane. To get a feeling for the molecule, follow the red strand around its cycle: It is linked twice to the green strand, twice to the cyan strand, twice to the magenta strand, and twice to the dark blue strand. It is only indirectly linked to the yellow strand. Note that each edge of the cube is a piece of double helical DNA, containing two turns of the double helix.
DNA Origami
Wyss Institute researchers are at the forefront of work in DNA Origami, a technique for folding pieces of DNA into shapes that may one day prove useful in manufacturing and medicine. The hope is that these incredibly tiny forms could carry cancer drugs deep inside the body or work as cogs in a molecular machine.
Recent advances by Wyss scientists include the formation of three-dimensional shapes, such as an icosahedron, and of twisted and curved shapes. It’s not yet clear which of these shapes will be most useful, but by developing expertise in shape creation and variety, scientists believe they will be able to make the best use of the technique.
The researchers’ short-term goal is to make increasingly complex shapes, improve their fabrication methods, and develop new applications for their nanoscale parts.
Long-term, Wyss Faculty Member William Shih recently told MSNBC. "We'd like to be able to approach the efficiency with which viruses deliver their cargo to cells, but do it in a safer way."
Link: http://wyss.harvard.edu/viewpage/199
Online DNA Assembly Simulator
Scientists at the Wyss Institute, led by Core Faculty members, William Shih and Peng Yin, have made available online a simulator of their work toward building complex, three-dimensional structures out of DNA. They call it, DNA origami.
With DNA origami, researchers take a long, single strand of DNA (called a scaffold) and fold it into a structure of their own design. These designs are held together with short "staple" strands, which are also made of DNA. Each staple is coded with two specific sequences: one is the complement to a specific site on the scaffold; the other is the complement to a second location on the same scaffold. When the scaffold and staples are allowed to "self-assemble," the attachment of the staple strands causes the scaffold to bend and fold into the desired structure. In principle, the same approach could be used with RNA or any other informational molecule where complementary sequences can be engineered.
Below is an interactive feature that illustrates the basic idea of DNA origami by letting you manipulate a biomolecule, such as DNA or RNA. The general idea is the same as DNA origami, but instead of adding staple strands to the scaffold, you define the binding sequences on scaffold, keeping in mind how you'd like the strand to fold. When you're satisfied with your coding, click on the "Self-Assemble" button and watch the strand fold up on itself, connecting at points you've selected. The molecule should take shape as planned -- or not! Either way, you might discover something interesting as you build.
Link: http://wyss.harvard.edu/viewpage/m-o/interactive-feature-molecular-origami
Links and References
Ned Seeman's Laboratory home page: http://seemanlab4.chem.nyu.edu/.